Carbohydrate Remediation
As the archive grows and science evolves, the 3D structures that are represented in the PDB Core Archive require ongoing improvement ("remediation") to ensure consistency, accuracy, and overall quality of the PDB archive. This will further the goals of wwPDB to make PDB data Findable, Accessible, Interoperable and Reusable (FAIR). While numerous tools exist to curate protein 3D structural data, few have been developed for consistent standard representation and for checking the veracity of carbohydrate 3D structural data within wwPDB deposition-validation-biocuration processes. Working with the glycoscience community, carbohydrate-appropriate annotation tools are being developed and implemented within the wwPDB OneDep unified system for deposition, validation, and biocuration of structures coming into the PDB Core Archive. These software tools will provide standard nomenclature and consistent oligosaccharide representation that can be easily translated to other representations commonly used by glycobiologists.
This effort will involve the following steps: (1) standardizing sugar nomenclature following IUPAC/IUBMB; (2) providing a uniform polymer representation for polysaccharides with appropriate descriptor(s); (3) adopting community software for reliable carbohydrate identification, assignment of standard nomenclature, and detection of intra/inter-molecular connectivity between monosaccharides and other molecules/proteins; and (4) providing intra- and inter-molecular connectivity at atom level explicitly. Due to the nature of carbohydrates, which are usually branched and/or have different glycosidic linkages, additional data descriptors will be introduced into the PDBx/mmCIF dictionary to describe new representation of oligosaccharides as branched entities. The branch components (monosaccharides) and their intramolecular linkages, as well as the glycosidic bonds, will be explicitly listed. The proposed PDBx/mmCIF dictionary extensions, some examples of how PDB Core Archive structures containing carbohydrates would be represented in PDBx/mmCIF format and the corresponding proposed Chemical Component Dictionary definitions are provided at github https://github.com/pdbxmmcifwg.
Each oligosaccharide molecule will be classified as a branched entity at entity.type data item, identified as oligosaccharide type at pdbx_entity_branch.type data item, and assigned a unique chain ID in _atom_site.auth_asym_id data item. The monosaccharides constituting an oligosaccharide entity will be listed in the pdbx_entity_branch_list category. Unlike polypeptides, where the peptidic bond is implicitly assumed, the connectivity of the glycosidic bond for an oligosaccharide entity is explicitly described at the entity level in pdbx_entity_branch_link category so that users can retrieve an oligosaccharide entity with correct linkages.
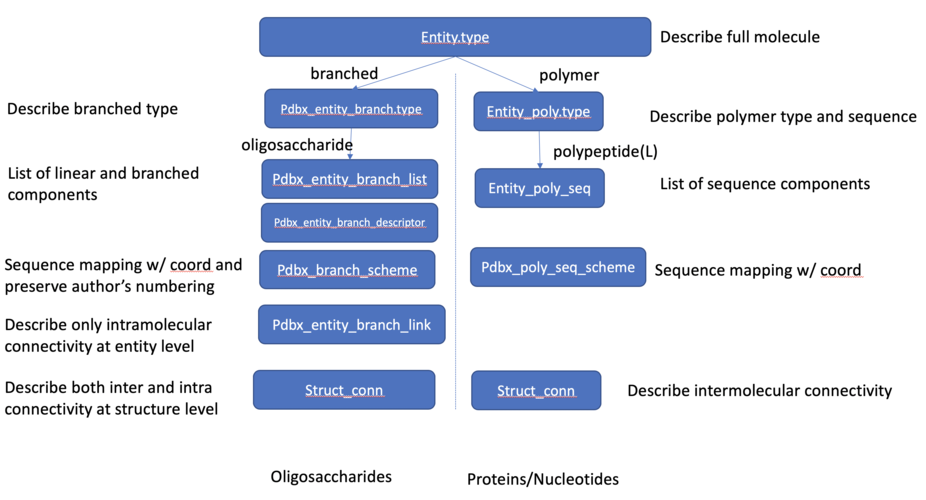
Figure 1. Oligosaccharide representation compared to protein/nucleic acid representation, branched vs polymer.
For example, the oligosaccharide, Lewis Y blood group antigen with components
(FUC)-NAG-GAL-FUC in PDB entry 2WMG will be represented as below branched entity.
#
_entry.id 2WMG
#
loop_
_entity.id
_entity.type
_entity.src_method
_entity.pdbx_description
_entity.formula_weight
_entity.pdbx_number_of_molecules
_entity.pdbx_ec
_entity.pdbx_mutation
_entity.pdbx_fragment
_entity.details
1 polymer man 'FUCOLECTIN-RELATED PROTEIN' 66769.758 1 ? YES 'CATALYTIC MODULE, RESIDUES 31-589' ?
2 branched nat "LFucpa1-2DGalpb1-4[LFucpa1-3]DGlcpNAcb" 681.7 1 ? ? ? ?
3 water nat water 18.015 334 ? ? ? ?
#
_pdbx_entity_branch.entity_id 2
_pdbx_entity_branch.type oligosaccharide
#
loop_
_pdbx_entity_branch_list.entity_id
_pdbx_entity_branch_list.comp_id
_pdbx_entity_branch_list.num
_pdbx_entity_branch_list.hetero
2 NAG 1 n
2 GAL 2 n
2 FUC 3 n
2 FUC 4 n
#
loop_
_pdbx_entity_branch_link.link_id
_pdbx_entity_branch_link.entity_id
_pdbx_entity_branch_link.entity_branch_list_num_1
_pdbx_entity_branch_link.comp_id_1
_pdbx_entity_branch_link.atom_id_1
_pdbx_entity_branch_link.leaving_atom_id_1
_pdbx_entity_branch_link.atom_stereo_config_1
_pdbx_entity_branch_link.entity_branch_list_num_2
_pdbx_entity_branch_link.comp_id_2
_pdbx_entity_branch_link.atom_id_2
_pdbx_entity_branch_link.leaving_atom_id_2
_pdbx_entity_branch_link.atom_stereo_config_2
_pdbx_entity_branch_link.value_order
_pdbx_entity_branch_link.details
1 2 1 NAG O4 HO4 ? 2 GAL C1 O1 R SING ?
2 2 2 GAL O2 HO4 ? 3 FUC C1 O1 R SING ?
3 2 1 NAG O3 HO3 ? 4 FUC C1 O1 R SING ?
#
To minimize impact to PDB Core Archive users, the current struct_conn that describes connectivity at structure level will remain as is, with extension to explicitly describe the leaving atom and its stereochemical configuration. (N.B.: Oligosaccharides are not represented in entity_poly_seq, hence sequential sequence numbering in _atom_site.label_seq_id will be null). Based on community feedback, the glycosylation site and its binding type (N-linked glycan or O-linked glycan) needs to be identified in a machine readable format in order to be easily parsed by users. The item struct_conn.pdbx_role is used for binding site and is a controlled vocabulary.
For example, the glycosylation sites are identified in _struct_conn.pdbx_role for PDB entry 4OF3.
#
#
_struct_conn.id
_struct_conn.conn_type_id
_struct_conn.pdbx_leaving_atom_flag
_struct_conn.ptnr1_label_comp_id
_struct_conn.ptnr1_label_asym_id
_struct_conn.ptnr1_label_seq_id
_struct_conn.ptnr1_label_atom_id
_struct_conn.pdbx_ptnr1_label_alt_id
_struct_conn.pdbx_ptnr1_PDB_ins_code
_struct_conn.pdbx_ptnr1_standard_comp_id
_struct_conn.pdbx_ptnr1_leaving_atom_id
_struct_conn.pdbx_ptnr1_atom_stereo_config
_struct_conn.ptnr1_symmetry
_struct_conn.ptnr2_label_comp_id
_struct_conn.ptnr2_label_asym_id
_struct_conn.ptnr2_label_seq_id
_struct_conn.ptnr2_label_atom_id
_struct_conn.pdbx_ptnr2_label_alt_id
_struct_conn.pdbx_ptnr2_PDB_ins_code
_struct_conn.pdbx_ptnr2_standard_comp_id
_struct_conn.pdbx_ptnr2_leaving_atom_id
_struct_conn.pdbx_ptnr2_atom_stereo_config
_struct_conn.ptnr2_symmetry
_struct_conn.pdbx_role
_struct_conn.pdbx_dist_value
_struct_conn.pdbx_value_order
_struct_conn.details
covale1 covale Y ASN A 93 ND2 ? ? ? OXT N 1_555 NAG C 801 C1 ? ? ? O1 R 1_555 N-Glycosylation 1.438 sing ?
covale2 covale Y ASN A 206 ND2 ? ? ? OXT N 1_555 NAG D 804 C1 ? ? ? O1 R 1_555 N-Glycosylation 1.415 sing ?
covale3 covale Y ASN B 93 ND2 ? ? ? OXT N 1_555 NAG E 801 C1 ? ? ? O1 R 1_555 N-Glycosylation 1.434 sing ?
#
The linear sequence descriptor of an oligosaccharide based on observed sequence will be described in _pdbx_entity_descriptor category so that it is interoperable with various representations that are used in the glycoscience community.
For example, the linear sequence descriptor for the oligosaccharide, (FUC)-NAG-GAL-FUC, is described in _pdbx_entity_descriptor.descriptor for PDB entry 2WMG.
#
loop_
_pdbx_entity_branch_descriptor.ordinal
_pdbx_entity_branch_descriptor.entity_id
_pdbx_entity_branch_descriptor.descriptor
_pdbx_entity_branch_descriptor.type
_pdbx_entity_branch_descriptor.program
_pdbx_entity_branch_descriptor.program_version
1 2 LFucpa1-2DGalpb1-4[LFucpa1-3]DGlcpNAcb1
"Glycam Condensed Sequence" GMML 1.0
2 2 [b-D-GlcpNAc]{[(3+1)][a-L-Fucp]{}[(4+1)][b-D-Galp]{[(2+1)][a-L-Fucp]{}}}
LINUCS PDB-CARE ?
#
In addition, we will standardize wwPDB Chemical Component Dictionary (CCD) so that all sugars are represented as monosaccharides with IUPAC style residue naming and standard atom nomenclature following IUPAC-IUBMB. For pyranoses, the leaving atom is O1, while for furanoses and sialic acid derivatives, the leaving atom is O2. These standardized atom nomenclatures will facilitate systematic numbering of components in oligosaccharides and also in validation of carbohydrate structures.
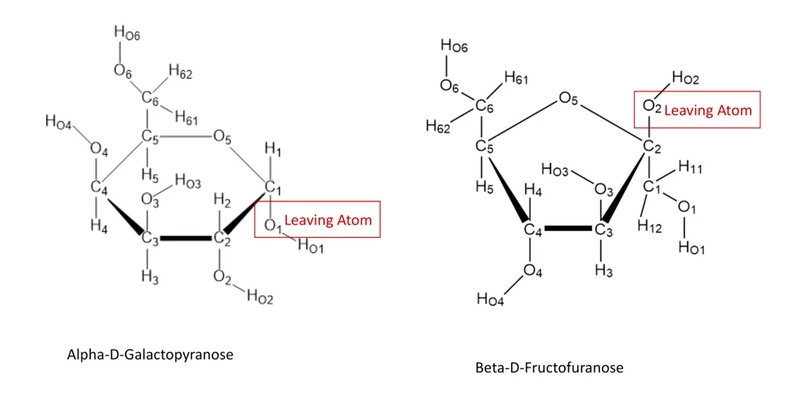
Figure 2. Standard atom nomenclature of pyranose and furanose ring sugars.
To support findability, wwPDB will preserve the commonly used names for oligosaccharides through extending an existing reference dictionary of biologically interesting molecules. To minimize the impact on the coordinate section (atom_site) of the PDB Core Archive files and on other software used in the field such as refinement programs or 3D visualization tools, the three letter code residue names will be retained in _atom_site.label_comp_id. We will introduce IUPAC style naming using existing data item, pdbx_chem_comp_identifier in the CCD for consistent annotation in the PDB Core Archive atomic coordinate files. This information will be copied to the PDB Core Archive atomic coordinate files as metadata.
For example, the IUPAC style naming b-D-GlcpNAc or GlcpNAcb or N-acetyl-b-D-glucopyranose-osamine for NAG are provided with source provenance identified.
#
loop_
_pdbx_chem_comp_identifier.comp_id
_pdbx_chem_comp_identifier.type
_pdbx_chem_comp_identifier.program
_pdbx_chem_comp_identifier.program_version
_pdbx_chem_comp_identifier.identifier
NAG "SYSTEMATIC NAME" ACDLabs 12.01
2-(acetylamino)-2-deoxy-beta-D-glucopyranose
NAG "SYSTEMATIC NAME" "OpenEye OEToolkits" 1.7.6
N-[(2R,3R,4R,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]ethanamide
NAG "CONDENSED IUPAC CARB SYMBOL" GMML 1.0
DGlcpNAcb
NAG "COMMON NAME" GMML 1.0
N-acetyl-b-D-glucopyranose-osamine
NAG "IUPAC CARB SYMBOL" PDB-CARE 1.0
b-D-GlcpNAc
NAG "SNFG CARB SYMBOL" GMML 1.0
GlcNAc
We will take this opportunity to improve management of synonyms in the wwPDB CCD during this remediation. We will move synonym list out of _chem_comp.pdbx_synonyms to a better organized table structure, pdbx_chem_comp_synonyms with source provenance.
For example, each synonym is listed at each row with data source identified for ligands ROC and for 58A.
#
loop_
_pdbx_chem_comp_synonyms.comp_id
_pdbx_chem_comp_synonyms.name
_pdbx_chem_comp_synonyms.provenance
_pdbx_chem_comp_synonyms.type
ROC Fortovase DRUGBANK ?
ROC SAQUINAVIR DRUGBANK ?
ROC "RO 31-8959" ? ?
#
loop_
_pdbx_chem_comp_synonyms.comp_id
_pdbx_chem_comp_synonyms.name
_pdbx_chem_comp_synonyms.provenance
_pdbx_chem_comp_synonyms.type
58A cytidinediphosphate-dioleoylglycerol PUBCHEM ?
58A CDP-1,2-dioleoyl-sn-glycerol CHEBI ?
#
#
Based on the feedback from glycoscience community, the relationship between a modified monosaccharide and its core structure (standard monosaccharide) is described in _pdbx_chem_comp_related category and the atom-to-atom mapping is provided in _pdbx_chem_comp_atom_related category.
For example, the core structure for modified monosaccharide, SGN(N-sulfo-6-O-sulfo-alpha-D-glucocosamine), is PA1(alpha-D-glucosamine) with "carbohydrate core" identified as relationship type.
#
_pdbx_chem_comp_related.comp_id SGN
_pdbx_chem_comp_related.related_comp_id PA1
_pdbx_chem_comp_related.relationship_type 'Carbohydrate core'
_pdbx_chem_comp_related.details ?
#
loop_
_pdbx_chem_comp_atom_related.ordinal
_pdbx_chem_comp_atom_related.comp_id
_pdbx_chem_comp_atom_related.atom_id
_pdbx_chem_comp_atom_related.related_comp_id
_pdbx_chem_comp_atom_related.related_atom_id
_pdbx_chem_comp_atom_related.related_type
1 SGN C1 PA1 C1 'Carbohydrate core'
2 SGN C2 PA1 C2 'Carbohydrate core'
3 SGN C3 PA1 C3 'Carbohydrate core'
4 SGN C4 PA1 C4 'Carbohydrate core'
5 SGN C5 PA1 C5 'Carbohydrate core'
6 SGN C6 PA1 C6 'Carbohydrate core'
7 SGN N2 PA1 N2 'Carbohydrate core'
8 SGN O1 PA1 O1 'Carbohydrate core'
9 SGN O3 PA1 O3 'Carbohydrate core'
10 SGN O4 PA1 O4 'Carbohydrate core'
11 SGN O5 PA1 O5 'Carbohydrate core'
12 SGN O6 PA1 O6 'Carbohydrate core'
21 SGN H1 PA1 H1 'Carbohydrate core'
22 SGN H2 PA1 H2 'Carbohydrate core'
23 SGN H3 PA1 H3 'Carbohydrate core'
24 SGN H4 PA1 H4 'Carbohydrate core'
25 SGN H5 PA1 H5 'Carbohydrate core'
26 SGN H61 PA1 H61 'Carbohydrate core'
27 SGN H62 PA1 H62 'Carbohydrate core'
28 SGN HN2 PA1 HN2 'Carbohydrate core'
29 SGN HO1 PA1 HO1 'Carbohydrate core'
30 SGN HO3 PA1 HO3 'Carbohydrate core'
31 SGN HO4 PA1 HO4 'Carbohydrate core'
#
Due to significant changes made to the CCD to support carbohydrate remediation, the enhanced CCD will be versioned and provided at versioned FTP (https://ftp-versioned.wwpdb.org) with better organization in the revision history which was already implemented in the atomic coordinate files. The main FTP archive will have the latest version of CCD, while every latest major version will be provided at versioned FTP archive.
For example, the revision history for a chemical component is captured in _pdbx_audit_revision_history and the existing mmCIF categories in the CCD that were updated during this remediation are listed in _pdbx_audit_revision_category.
#
loop_
_pdbx_audit_revision_history.ordinal
_pdbx_audit_revision_history.data_content_type
_pdbx_audit_revision_history.major_revision
_pdbx_audit_revision_history.minor_revision
_pdbx_audit_revision_history.revision_date
1 'Chemical component' 1 0 1999-07-08
2 'Chemical component' 1 1 2011-06-04
3 'Chemical component' 2 0 2018-10-17
#
loop_
_pdbx_audit_revision_details.ordinal
_pdbx_audit_revision_details.revision_ordinal
_pdbx_audit_revision_details.data_content_type
_pdbx_audit_revision_details.provider
_pdbx_audit_revision_details.type
_pdbx_audit_revision_details.description
1 1 'Chemical component' repository 'Initial release' ?
#
loop_
_pdbx_audit_revision_category.ordinal
_pdbx_audit_revision_category.revision_ordinal
_pdbx_audit_revision_category.data_content_type
_pdbx_audit_revision_category.category
1 3 'Chemical component' '_chem_comp'
2 3 'Chemical component' '_chem_comp_atom'
3 3 'Chemical component' '_pdbx_chem_comp_synonyms'
4 3 'Chemical component' '_pdbx_chem_comp_identifier'
#
We plan the rollout of new representation of carbohydrate structures to the PDB Core Archive in the early 2020. The data representation is a result of consultation with glycoscience community and the wwPDB PDBx/mmCIF Working Group via discussions with key stakeholders at annual carbohydrate/glycobiology and recurring Working Group meetings. A more substantial collection of example files of remediated data representing various use cases will be provided for testing and adoption by the glycoscience community, refinement software developers, cheminformaticians, and 3D visualization key stakeholders during the development stage.
Acknowledgements
The carbohydrate remediation project is a wwPDB collaborative project that is carried out principally by RCSB PDB at Rutgers, The State University of New Jersey and is funded by NIGMS grant U01 CA221216 in collaboration with Complex Carbohydrate Research Center at the University of Georgia.