wwPDB 2025 News
Contents
09/23/2025
Introducing the first 3DEM Model-Map percentile slider to the wwPDB validation report
The wwPDB is continuing to improve and expand our validation offerings with a new addition to the wwPDB PDF validation reports. Building on the success of the coordinate‑model percentile sliders, which help depositors, reviewers, and archive users assess PDB model quality in the context of the PDB archive, we are now extending this section to include a Q-score percentile slider. The new slider is the first to empower users of this report to assess model-map quality at a glance relative to the EMDB/PDB archives.
Starting October 01, newly released 3DEM entries will include the Q‑score percentile slider in its own section below the current sliders under “Overall quality at a glance.” The slider compares an entry’s average Q‑score against both the entire EMDB/PDB archive and a resolution‑similar subset. Because Q‑score correlates strongly with resolution between 1–10 Å, the percentile helps check whether a reported global resolution is reasonable; unusually low values can flag model–map fit or map quality issues. A dedicated Q‑score section supports deeper review with residue‑level mapping to the coordinate model and per‑chain tables.
In the near future, wwPDB will re-generate validation reports with Q-score slider for all existing entries in the PDB archive.
We are excited for our users to get their hands on this new validation metric. If you have any queries or feedback please let us know at info@wwpdb.org.
 3DEM Model-Map percentile slider from the wwPDB validation report
3DEM Model-Map percentile slider from the wwPDB validation report
09/22/2025
MolViewSpec: describe, share, and reproduce Mol* molecular scenes
 Graphical abstract from MolViewSpec: a Mol* extension for describing and sharing molecular visualizations
Graphical abstract from MolViewSpec: a Mol* extension for describing and sharing molecular visualizationsMolViewSpec is a new Mol* extension to create molecular scenes that allows users to:
- Describe molecular scenes: what to load (structures, maps, annotations) and how to show it (representations, colours, labels, measurements, custom 3D shapes…). You can mix data types (structural, volumetric, annotations) in one scene with consistent styling.
- Share: to use the scenes, simply open the spec in Mol* in the browser or use the standalone Python package for scripted workflows.
- Reproduce: scenes make visuals reproducible across browsers, collaborators, and notebooks. No more “how did you make that figure?”.
- Automate figure creation and analysis by generating specs from Python pipelines.
Learn more and try examples at molstar.org
Read more:
MolViewSpec: a Mol* extension for describing and sharing molecular visualizations
Adam Midlik, Sebastian Bittrich, Jennifer R Fleming, Sreenath Nair, Sameer Velankar, Stephen K Burley, Jasmine Y Young, Brinda Vallat, David Sehnal
(2025) Nucleic Acids Research 53: W408–W414 10.1093/nar/gkaf370
08/18/2025
Paper Published: 3DEM Structure-Map Validation Recommendations
A new publication describes two percentile-based metrics: Q_relative_all and Q_relative_resolution. The publication presents a statistical analysis of Q-score and these new percentile based validation metrics.
Q_relative_all is a single value percentile, representing the overall quality of the model-map fitness. It is derived by comparing a model-map average Q-score to all model-map average Q-scores calculated for 3DEM structures in the PDB/EMDB. Higher Q_relative_all percentile values, infer higher quality in the model-map fitness.
Q_relative_resolution is a single value percentile, derived by comparing the average Q-score of a model-map with the Q-scores of other model-map entries of similar resolution. This metric was developed from the principle that average Q-score is correlated with map resolution. Q_relative_resolution percentile values close to 50% therefore represent a score that is commonly observed for entries in the PDB/EMDB of similar resolution, inferring model-map fitness is typical for an entry of a particular resolution. Notably low or high Q_relative_resolution percentiles may be helpful to guide further review of model-maps.
The EMDB has made these metrics available since Spring 2024 via emdatabank.org and since the publication of the paper, the wwPDB is in the process of incorporating these metrics into wwPDB validation reports by providing the Q-score percentile slider for the assessment of model-map fitness of 3DEM entries. This validation enhancement will help depositors, reviewers and the community to improve the quality of data submissions and assist users to assess 3DEM data quality in the PDB and EMDB, wwPDB core archives.
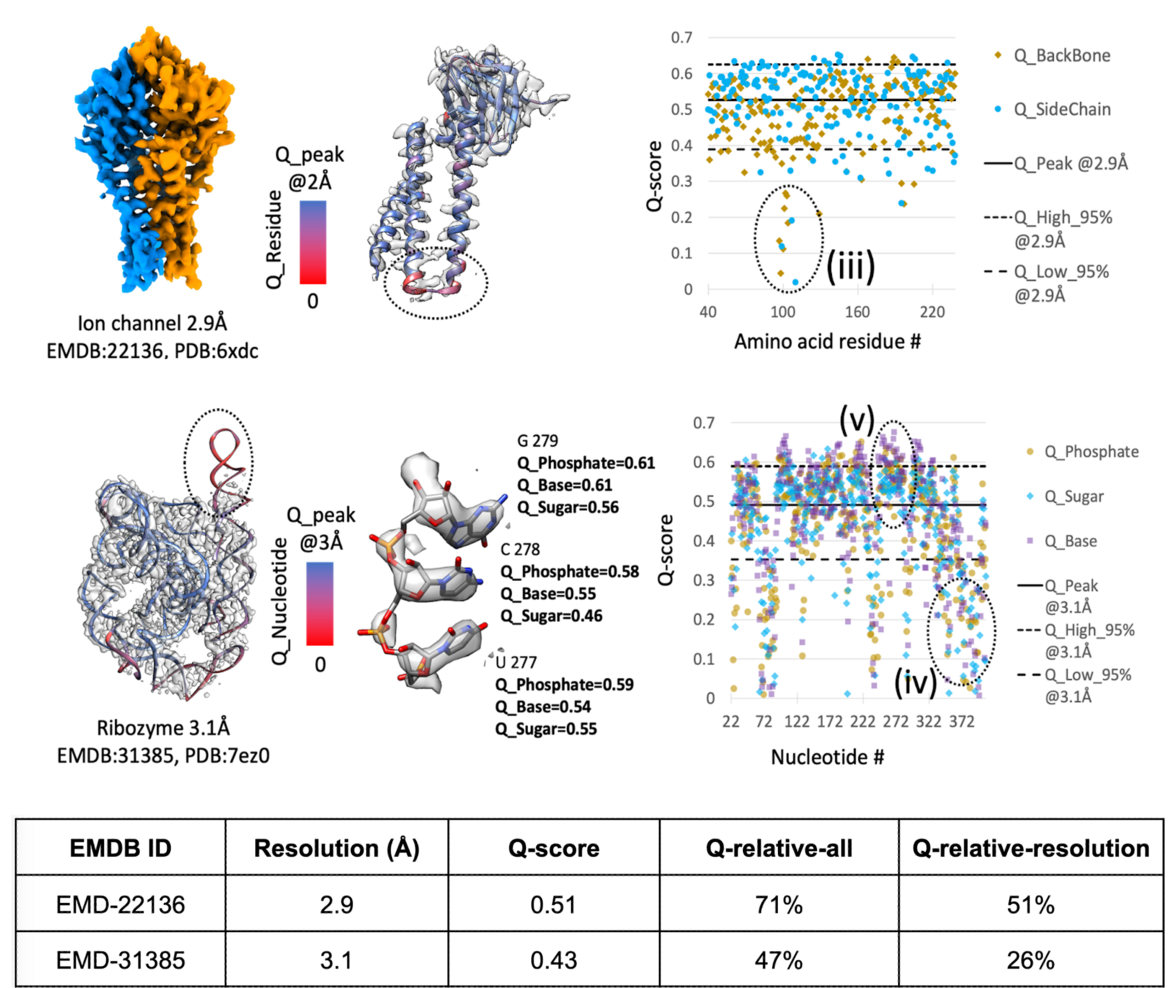 Examples of Q-score application in proteins and in nucleic acids adapted from: doi: 10.1107/S2059798325005923
Examples of Q-score application in proteins and in nucleic acids adapted from: doi: 10.1107/S2059798325005923 Q-score as a reliability measure for protein, nucleic acid and small-molecule atomic coordinate models derived from 3DEM maps
Grigore Pintilie, Chenghua Shao, Zhe Wang, Brian P Hudson, Justin W Flatt, Michael F Schmid, Kyle L Morris, Stephen K Burley, Wah Chiu
(2025) Acta Cryst D81: 410-422 doi: 10.1107/S2059798325005923
07/15/2025
Transitioning to PDBx/mmCIF and Extended PDB IDs
wwPDB strongly encourages all users to adopt the extended PDB ID format and transition to PDBx/mmCIF file format as soon as possible. This includes making changes to software; referring to structures by the full 12-character ID in all communications; and encouraging your communities to do the same.
Transitioning to Extended PDB IDs
As the PDB archive continues to expand, the four-character PDB accession codes (PDB IDs) are expected to be fully assigned before 2028. To support the growth of the archive, the wwPDB has extended the length of PDB IDs to 12 alphanumeric characters including "pdb_" prefix (e.g., "1abc" will become "pdb_00001abc", case insensitive) to improve text mining capabilities in the published literature. Users or journals will be able to parse/recognize PDB IDs using the prefix “pdb_”. The prefix and zeros must be included in the extended PDB ID.
Once four-character PDB IDs are fully assigned, new entries will only receive extended PDB IDs; data will not be provided in the legacy PDB file format files.
Access further details, including a transition plan, example files, and supporting FAQs at wwPDB: Extended PDB ID With 12 Characters.
Users can adopt usage of extended PDB IDs for all PDB entries immediately using the _database_2.pdbx_database_accession data item in the PDBx/mmCIF formatted structure files.
For example:
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB 2HYV pdb_00002hyv 10.2210/pdb2hyv/pdb
WWPDB D_1000038924 ? ?
New PDB DOI Format
All existing PDB entries with four-character PDB IDs issued have DOI formatted as 10.2210/pdb[4-character_PDB_ID]/pdb that resolve to the corresponding wwPDB DOI landing page. For example, PDB entry 8y9m (pdb_00008y9m) has the DOI https://doi.org/10.2210/pdb8y9m/pdb. Importantly, this DOI will remain unchanged in the future.
When all 4-character PDB IDs have been exhausted, all new PDB entries will be issued extended PDB IDs issued and a NEW DOI formatted as 10.2210/[Extended_PDB_ID]/pdb that will resolve to the corresponding wwPDB DOI landing page. For example, PDB entry “pdb_10001xyz” will have the DOI https://doi.org/10.2210/pdb_10001xyz/pdb.
Transitioning to PDBx/mmCIF Format
To help users adopt extended PDB ID and PDBx/mmCIF file format, wwPDB offers an mmCIF User Guide and software resources such as mmCIF parsers and CIF Editor.
In addition, wwPDB will provide a Beta PDB Archive organized by extended PDB ID (including file naming, directories, and datablock naming) in early 2026. The current PDB archive organizes data files grouped by data type, e.g., coordinates, experimental data, assemblies, validation reports, etc.
A major change in the Beta PDB archive will be the re-organization of file directory at entry level, following the same file organization as the PDB Versioned Archive. In other words, all the data files associated to an entry will be grouped together under its PDB ID (extended PDB ID) with two letter hash. Please watch wwPDB.org and community bulletin boards for announcements on the file organization for the Beta PDB archive later this year.
We recommend users fully adopt all of these these changes before the end of 2026. Early adoption will contribute to the long-term sustainability and interoperability of 3D biostructure data across the scientific community.
In particular, journals should begin adopting the Extended PDB ID format (in Text, Tables, and Data Availability Statements), updating links included in journal articles for PDB IDs to the wwPDB DOI landing page via CrossRef, and verifying software tools linked from a journal article (e.g., FirstGlance or Jsmol for 3D visualization) support of extended PDB IDs and PDBx/mmCIF.
Should you have any questions or require further assistance, please do not hesitate to contact us at info@wwpdb.org. We greatly appreciate your support and cooperation as we work together to enhance the future of structural data accessibility.
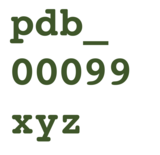 Sample extended PDB IDSample extended PDB ID
Sample extended PDB IDSample extended PDB ID
06/25/2025
Extension to EMDB Accession Codes
The EMDB archive is experiencing near-exponential growth. To support this rapid expansion, the wwPDB has been working to prepare our systems for the future and ensure seamless deposition, validation, and dissemination of data in the years ahead.
As part of this effort, we are announcing an important change:
EMDB IDs will be extended to support up to six digits, allowing for identifiers such as EMD-123456. Currently, the archive uses four and five-digit IDs (e.g., EMD-0123, EMD-45678), with support for identifiers up to the EMDB ID: EMD-99999. The change to six-digits will increase our capacity ten-fold and ensure ID availability into the next decade, up to EMD-999999.
We do not anticipate surpassing 99,999 released entries until around 2028, however, we expect to begin assigning six-digit EMDB IDs sometime in 2026. As such, as early as 2026, depositors should anticipate being assigned six-digit IDs within the wwPDB OneDep deposition system, and the user and developer community can expect six-digit EMDB IDs in the EMDB archive.
We appreciate your support as the archive grows and thank you for your contribution to the worldwide effort to archive structural biology data. We remain committed to ensuring a stable, sustainable and scalable infrastructure that supports the continued successes and productivity of the scientific community using structural biology and cryoEM in their research. For questions or feedback, contact the EMDB team at emdbhelp@ebi.ac.uk.
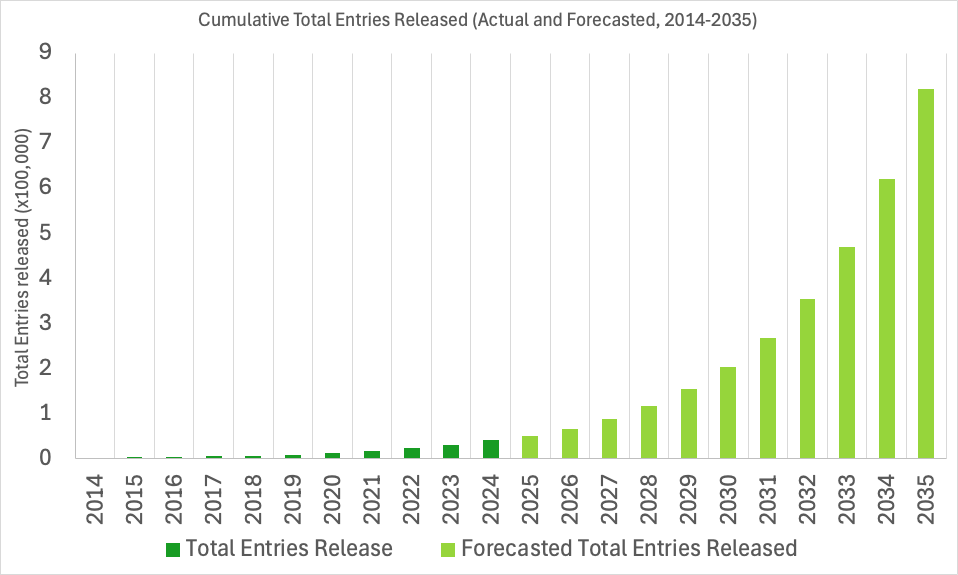 EMDB Growth
EMDB Growth
05/06/2025
Poster Prize Awarded at ASBMB
The wwPDB Foundation made an award for outstanding student presentation at the 2025 meeting of The American Society for Biochemistry and Molecular Biology (April 12–15; Chicago, IL).
 Maria Ahmed (University of Rochester) and Christine Zardecki (wwPDB Foundation)
Maria Ahmed (University of Rochester) and Christine Zardecki (wwPDB Foundation)Incorporation of methionine sulfoxides into nascent proteins during translation
Maria Ahmed, Philip Bellomio, Michael Meadow, Kevin Welle, Kyle Swovik, Jenny Hryhorenko, Sina Ghaemmaghami, University of Rochester
Many thanks to The Biophysical Society organizers and poster prize judges for making this award possible.
The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB50 events, workshops, and educational publications. The Foundation is chartered as a 501(c)(3) entity exclusively for scientific, literary, charitable, and educational purposes.
The wwPDB Foundation is grateful for our industrial sponsors: Discngine and MiTeGen. Individual sponsorships are also available.
Consider supporting the next 50 years of PDB's spirit of openness, cooperation, and education with a donation to the wwPDB Foundation.
04/16/2025
Download Snapshots of the PDB Core Archive via HTTPS, SYNC, RSYNC, or FTP
Snapshots of the PDB Core archive (https://files.rcsb.org) can be downloaded via HTTPS, SYNC, RSYNC, or FTP. At RCSB PDB, AWS SYNC is supported instead of RSYNC.
Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
HTTPS Protocol
AWS SYNC
RSYNC Protocol
- PDBj (Japan): rsync -avy snapshots.pdbj.org:: .
FTP Protocol
- PDBj (Japan): ftp://snapshots.pdbj.org
03/11/2025
Poster Prize Awarded at The Biophysical Society Meeting
The wwPDB Foundation made an award for an outstanding student presentation at the 2025 Biophysical Society Meeting (February 15-19, Los Angeles, CA).
 Hsiang-Ling Huang
Hsiang-Ling HuangMechanisms of dysferlin-mediated membrane repair in health and disease
Hsiang-Ling Huang (1), Giovanna Grandinetti (1,2), Sarah M. Heissler (1), Krishna Chinthalapudi (1)
(1) Department of Physiology and Cell Biology, Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University College of Medicine, United States
(2) Center for Electron Microscopy and Analysis, The Ohio State University, United States
Many thanks to The Biophysical Society organizers and poster prize judges for making this award possible.
The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB50 events, workshops, and educational publications. The Foundation is chartered as a 501(c)(3) entity exclusively for scientific, literary, charitable, and educational purposes.
The wwPDB Foundation is grateful for our industrial sponsors: Discngine. Individual sponsorships are also available.
Consider supporting the next 50 years of PDB's spirit of openness, cooperation, and education with a donation to the wwPDB Foundation.
03/10/2025
New version of PDB-IHM validation reports now include assessments of crosslinking mass spectrometry-based structure models
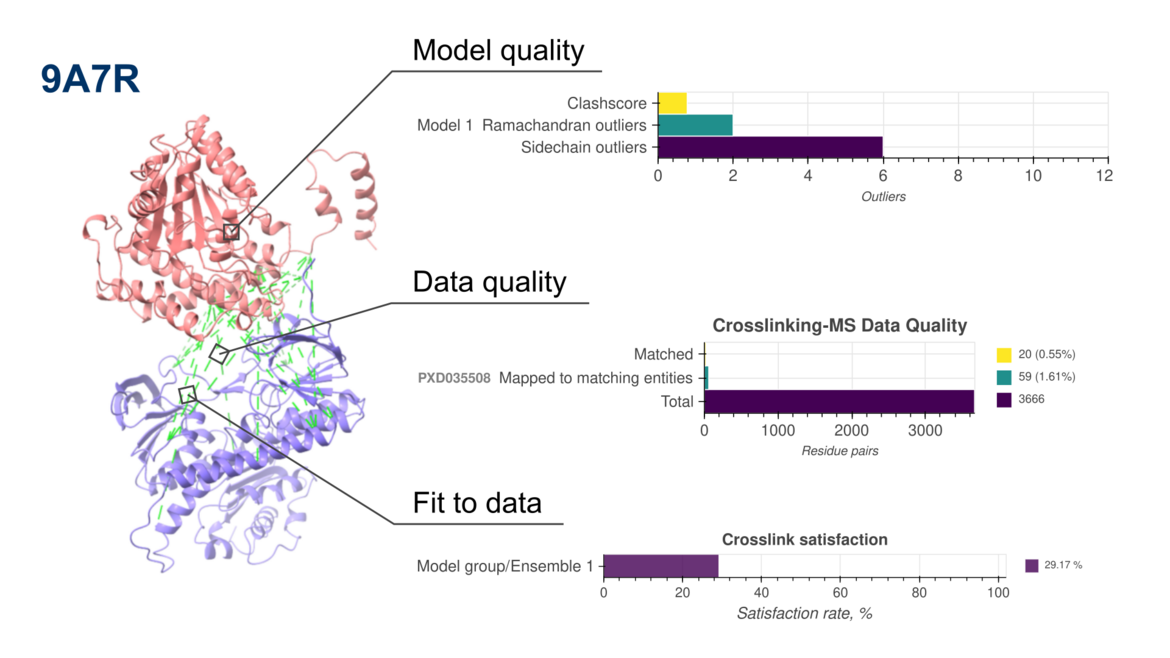 New version of PDB-IHM validation reports now include assessments of crosslinking mass spectrometry-based structure models
New version of PDB-IHM validation reports now include assessments of crosslinking mass spectrometry-based structure modelsWe are pleased to announce the release of updated validation reports (version 2) for structures determined using integrative and hybrid methods (IHM) archived in the PDB-IHM branch of the PDB archive. These reports are available in the archive and on the PDB-IHM website. IHM Validation software v2 is now integrated into the PDB-IHM deposition and curation system to support validation of new depositions. For detailed information on IHM Validation and v2 updates, please refer to the help page and user guide. The software is also available for standalone use via GitHub.
We thank the wwPDB IHM Task Force members and the broader structural biology community for supporting the development of validation methods for integrative structures.
What’s New in Version 2?
The latest update extends IHM validation to structures derived from Chemical Crosslinking Mass Spectrometry (Crosslinking-MS) data, in addition to the previously supported Small Angle Scattering (SAS) data. This milestone is the result of close collaboration among the Crosslinking-MS community, PRIDE repository, and the PDB-IHM team.
To support this advancement, we have developed interoperability mechanisms between PDB-IHM and PRIDE, enabling validation of integrative structures based on Crosslinking-MS data. Additionally, PRIDE Crosslinking now supports submission of complete Crosslinking-MS datasets following the mzIdentML 1.2.0 community data standard.
Furthermore, IHM Validation software v2 uses MolProbity version 4.5.2 and the PrISM software for precision analysis.
Next Steps for Depositors
We encourage depositors working with Crosslinking-MS-based integrative structures to:
- Submit complete datasets to PRIDE Crosslinking following the provided guidelines.
- Cross-reference PRIDE Crosslinking datasets in PDB-IHM depositions to enable robust validation.
01/29/2025
Take the PDBx/mmCIF User Guide Survey and Win!
Please take this brief survey about the PDBx/mmCIF User Guide to help us gain insights into the Guide’s usage and identify areas for future improvement.
 Benefits of the PDBx/mmCIF ecosystem
Benefits of the PDBx/mmCIF ecosystemYour feedback is highly valued.
Respondents can enter a drawing for a chance to win a prize from the wwPDB.
The survey will remain open until February 28, 2025.
01/16/2025
PDB-Dev now PDB-IHM
 Integrative structures are available at wwPDB.org and the PDB archive
Integrative structures are available at wwPDB.org and the PDB archiveStructures of many large macromolecular assemblies are now being determined using integrative approaches, wherein information derived from multiple experimental and computational methods is combined to compute their three-dimensional structures. PDB-IHM (formerly PDB-Dev) is a system for archiving and disseminating structures determined using integrative or hybrid methods (IHM), and making them Findable, Accessible, Interoperable, and Reusable (FAIR).
In August 2024, PDB-Dev was unified with the PDB to deliver integrative structures alongside experimental structures in the PDB archive. With unification, integrative structures are assigned PDB accession codes and Digital Object Identifiers (DOIs), annotated as IHM structures, and can be accessed from the PDB archive, PDB DOI links (e.g., DOI: 10.2210/pdb8zzc/pdb), and the PDB-IHM website. Now part of the PDB infrastructure, PDB-Dev has been rebranded as PDB-IHM, denoting IHM structures archived in the PDB.
Integrative structures can be deposited through the PDB-IHM deposition portal and accessible from the wwPDB OneDep home page. They are processed in parallel to the wwPDB OneDep system. Structures processed by PDB-IHM are released synchronously with PDB structures weekly on Wednesdays at 00:00 UTC.
In the future, the wwPDB partners, including Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) in the United States, Protein Data Bank in Europe (PDBe), and Protein Data Bank Japan (PDBj), will disseminate integrative structures on their respective websites.
We look forward to supporting the structural biology community with depositing integrative structures to PDB-IHM.
Questions or feedback? Contact deposit-help@mail.wwpdb.org or heldesk@pdb-ihm.org.
01/05/2025
Time-stamped Copies of PDB and EMDB Archives
 New archive snapshots are available.
New archive snapshots are available. A snapshot of the PDB Core archive (https://files.wwpdb.org/, https://s3.rcsb.org) as of January 1, 2025 has been added to https://s3snapshots.rcsb.org/, snapshots.rcsb.org (rsync -rlpy -a -v --delete snapshots.rcsb.org:: .), and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20250101 includes the 229,564 experimentally-determined structure and experimental data available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot of PDB Core Archive is 1,437 GB.
A snapshot of the EMDB Core archive (ftp://ftp.ebi.ac.uk/pub/databases/emdb/) as of January 01, 2025 can be found in ftp://ftp.ebi.ac.uk/pub/databases/emdb_vault/20250101/ and ftp://snapshots.pdbj.org/20250101/. The snapshot of EMDB Core Archive contains map files and their metadata within XML files for both released and obsoleted entries (41,367 and 301, respectively) and is 21 TB in size.