wwPDB 2019 News
Contents
11/04/2019
Additional EM map validation now available through OneDep
The wwPDB validation reports provided in the OneDep system now have additional validation for electron microscopy (EM) maps to help users identify potential discrepancies in their data.
The updated wwPDB validation reports in the OneDep system now incorporate an extensive EM map validation process, integrating a range of established validation methods for EM data previously available on the EMDB pages. Initially, this additional EM validation is only provided to depositors in the OneDep system, however in future will be provided for entries throughout the PDB and EMDB archives.
The process includes an analysis of the fit of the PDB model to the EMDB map, represented at an amino acid level on the residue-property plots and globally by a visual overlay of the map and model (see below images). FSC curves are also included to compare reported and estimated resolution, where either half maps or FSC data was uploaded.
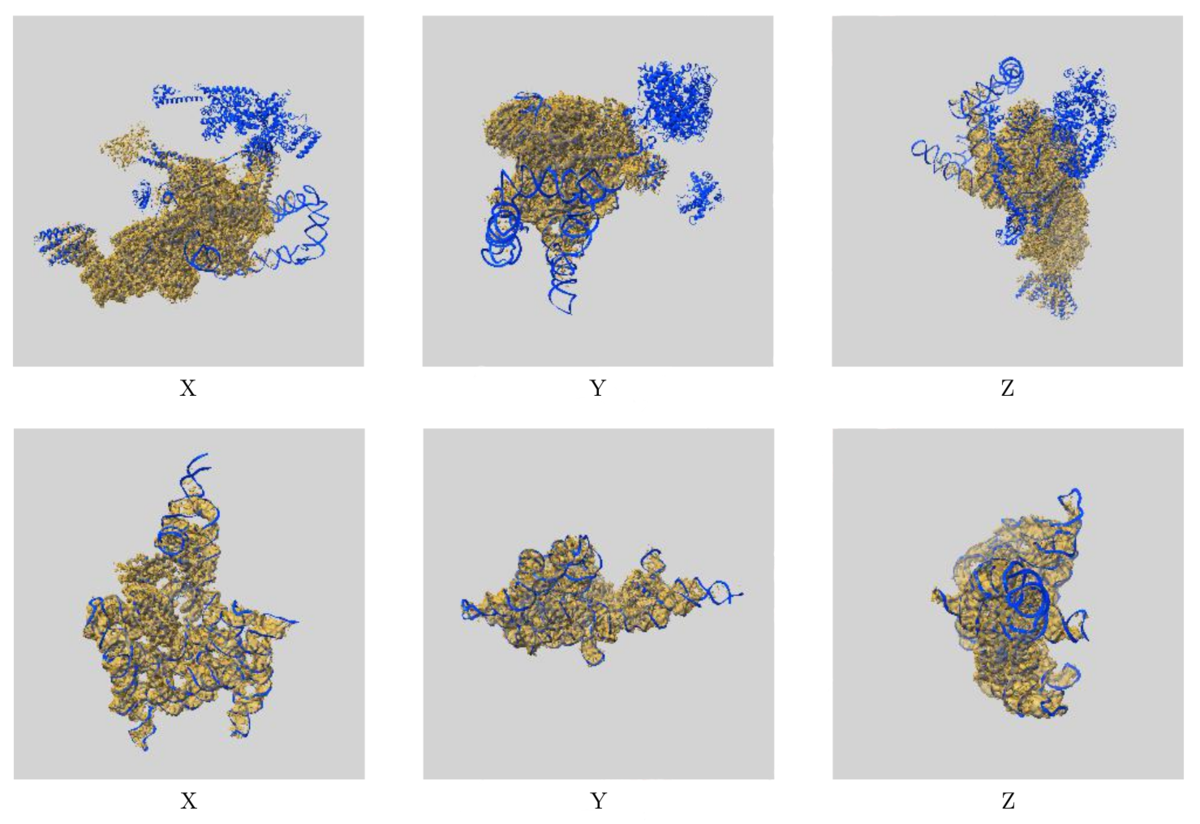 Images showing the visual EM map/model fit as displayed in the new validation reports. The top images display three orthogonal views of the map and model for EMD-0360 and 6N7P and highlights where there are regions of the model not covered by the EM map at the provided contour level, while the below images show the fit of EMD-9105 and 6ME0, where the majority of the model fits well to the map at the provided contour level.
Images showing the visual EM map/model fit as displayed in the new validation reports. The top images display three orthogonal views of the map and model for EMD-0360 and 6N7P and highlights where there are regions of the model not covered by the EM map at the provided contour level, while the below images show the fit of EMD-9105 and 6ME0, where the majority of the model fits well to the map at the provided contour level.The additional EM validation also includes various graphics for visual inspection of the data. Included in the reports are images of orthogonal projections, central slices, mask visualisation, and more, allowing for inspection of details in the map and identification of artifacts. A statistical analysis of the EM map volume is also provided, including graphs of map density distribution, volume estimate by contour, and rotationally averaged power spectrum, providing more thorough analysis of the EM volume.
These changes should help both depositors and users to identify potential errors in EM data and give more clarity about potential limitations of the data in both the PDB and EMDB.
Additional information about validation reports for EM entries is available.
If you have any questions or queries about wwPDB validation, then please contact us at validation@mail.wwpdb.org.
10/08/2019
Improved resolution of DOIs for PDB entries
The wwPDB partners are pleased to announce an improved mechanism for the resolution of digital object identifiers (DOIs) associated with released PDB entries. We have launched new wwPDB landing web-pages for each released PDB entry. These pages present basic information about the corresponding PDB structure, offer the model coordinate, experimental data and validation file downloads from the wwPDB FTP area, and, importantly, provide links to all the wwPDB partner websites that serve further advanced information about PDB structures. For an example, please navigate to the landing page for one of recently released entries in the PDB archive https://doi.org/10.2210/pdb6qw9/pdb. This development represents a significant improvement over the previous mechanism, where DOIs would resolve to a PDB-formatted file.
We encourage the scientific journals to make use of these pages and the DOIs issued for each PDB entry by linking to them from the online versions of papers where PDB entries are described or mentioned. We have also taken this opportunity to update the metadata that is associated with each PDB DOI, so that this information can be mined directly from the API offered by our DOI provider (CrossRef).
We would also like to draw the scientific community's attention to the new style of PDB accessions that are being gradually introduced, in preparation for when the supply of the familiar four character codes will be exhausted. The new accessions start with a prefix "PDB_" and contain further eight alphanumeric characters, with the last four characters identical to the familiar four-character codes.
07/31/2019
Improve your previously released coordinates AND keep your original PDB ID with OneDep
We are pleased to announce the availability of PDB versioning, allowing depositors to update their entries while retaining the same PDB accession code.
Depositors can now submit new coordinates for existing entries. Initially, this is limited to PDB entries that were submitted via the OneDep system, which was introduced in 2014. We plan to extend this functionality to entries deposited in the legacy systems (ADIT and Autodep) in future, and will announce a timeline for this in due course.
Requests should be initiated using the OneDep communication panel within the deposition session for the entry in question. Once submitted, the revised model will be processed by wwPDB biocurators and a new version released. Versioning of PDB entries will be limited to changes in the coordinate files, with no changes permitted to the deposited experimental data. PDB versioning will be limited to one replacement per PDB entry per year, and three entries per Principal Investigator per year.
The most recent version of the entry will be available in the main PDB archive FTP (http://ftp.wwpdb.org). All major versions of a PDB structure will be retained in the versioned FTP archive (ftp-versioned.wwpdb.org) - more information can be found on the wwPDB website. The structure of the versioned FTP archive has been built allowing for future extension of the PDB code format. PDB entry 1abc would therefore be found in the folder pdb_00001abc.
Changes made to entries during versioning are considered to be either “major” or “minor”. Updates to atomic coordinates, polymer sequence, or chemical description trigger a major version increment, while changes to any other categories are indicated as “minor”. Changes introduced are recorded in the PDBx/mmCIF audit categories.
If you have any further queries regarding the process of PDB versioning, please contact the wwPDB at deposit-help@mail.wwpdb.org.
06/30/2019
Mandatory PDBx/mmCIF format file submission for MX depositions
From today, July 1st 2019, submission of PDBx/mmCIF format files for crystallographic depositions to the PDB is mandatory.
PDBx/mmCIF is now the only format accepted for deposition of PDB structures resulting from macromolecular crystallography (MX), including X-ray, neutron, fiber, and electron diffraction methods via OneDep. The deposition of PDBx/mmCIF format files will improve the efficiency of the deposition process and enhance validation through capture of the more extensive experimental metadata supported by PDBx/mmCIF, compared to the legacy PDB format. PDB entries with 100,000 or more atoms, and those with multiple character chain IDs had no longer been supported by the legacy PDB format. In addition, by 2021, we anticipate the PDB Chemical Component Identifier will need to be extended beyond three characters, which will necessarily result in full retirement of files in the PDB Core Archive that utilize the legacy PDB format.
Refmac, Phenix.refine, and Buster programs can now output PDBx/mmCIF formatted files. For users of other structure determination/refinement software packages, the wwPDB provides stand-alone and web-based tools to convert legacy PDB format files into PDBx/mmCIF format: pdb_extract and MAXIT. More information on outputting and preparing PDBx/mmCIF format files for deposition can be found on the wwPDB website.
The PDBx/mmCIF Working Group has committed to the PDBx/mmCIF data model. PDBx/mmCIF is also supported by visualization software applications, including Jmol/JSMol, Chimera, OpenRasMol, CCP4MG, COOT, PyMOL, VMD, Molmil, LiteMol, Mol* and NGL. In addition, other data resources, such as the Protein Model Portal and SASBDB, have adopted and extended the PDBx/mmCIF framework for data representation.
If you have any further questions regarding deposition please contact the wwPDB on deposit-help@mail.wwpdb.org.
06/11/2019
Improvements to visualization of ligand validation and electron density maps in the wwPDB validation report
Our recent update to the wwPDB validation reports provides much clearer validation information for ligands.
We now include 2-dimensional diagrams of ligands, highlighting geometric validation criteria and, for structures determined by crystallography, 3-dimensional views of electron density.
We also provide calculated electron density map coefficients which were used to generate the analysis in the validation reports.
Ligand Validation
We have collaborated with Global Phasing Ltd to integrate the ligand visualization from buster-report into the wwPDB validation report, as recommended by the wwPDB/CCDC/D3R Ligand Validation Workshop. The ligand visualization will be available for ligands that have been designated as "Ligand of Interest" by the depositor and ligands with a molecular weight greater than 250 Daltons that have outliers.
The following ligand instance of NAP was chosen intentionally as a representative of sub-optimal quality in both the ligand model and its agreement with the X-ray data.
Geometric analysis provided by CCDC Mogul will be highlighted on a 2D diagram of the ligand, as shown below.
In addition to geometric validation for ligands, for X-ray diffraction PDB entries the wwPDB validation report also presents images displaying the ligand and the surrounding electron density map.
Electron Density Map Coefficient Files
We are now providing depositors with electron density map coefficient files (2mFo-DFc and mFo-DFc) from the wwPDB validation pipeline alongside the wwPDB validation report. The electron density map coefficients generated for wwPDB validation reports will be made available to end users in the PDB archive as new entries are released and for existing entries when validation reports for the PDB archive are recalculated.
We hope that these changes to the wwPDB validation pipeline will help depositors to interpret the validation information provided for PDB entries more easily. If you have any queries, please contact the wwPDB at deposit-help@mail.wwpdb.org.
06/03/2019
A tribute to Prof. Michael G. Rossmann
All of us at wwPDB were deeply saddened to hear of the recent passing of Prof. Michael G. Rossmann and our condolences go out to his family and friends at this time. Michael contributed so much to the field of structural biology and was a great supporter of the wwPDB and our activities. Here we would like to share our own tribute to Michael and his contribution to the PDB archiving efforts.
Michael has had an esteemed structural biology career, with over 300 structures to his name in the PDB, spanning from 1977 through to 2019. He also helped to develop the molecular replacement technique, enabling phasing of data in X-ray crystallography, which has been used for structure determination of around 100,000 PDB entries to date. One of Michael's key discoveries is of the Rossmann fold, a structural motif commonly found in enzymes that bind to dinucleotide cofactors. It's importance is highlighted by the fact that around 20,000 structures in the PDB contain a Rossmann fold motif, another lasting legacy of his work within the PDB archive.
Michael also made huge contributions to the field of virology, with hundreds of his virus structures archived in the PDB. This includes the first structure determined of the human rhinovirus, otherwise known as the common cold. His work has significantly improved the knowledge of virus structure and function, supporting development of new therapeutics to treat and prevent viral infections.
Michael was a member of the wwPDB advisory committee for four years, serving on the board from 2009 to 2012, and was also involved in advisory committees for RCSB PDB and EMDataBank. During this time, he provided much advice and guidance to the wwPDB, helping to shape the archiving activities during this time and into the future. Helen Berman, former director of the RCSB Protein Data Bank, highlights how Michael "was an active supporter of the Protein Data Bank from its inception in the early 1970s", while Haruki Nakamura, former head of Protein Data Bank Japan (PDBj), speaks of "always being impressed by his thoughtful comments about the activities of the wwPDB." Haruki adds: "Michael understood the importance of archiving raw experimental data and he had promoted 'data science' for many years."
Gerard Kleywegt, former head of Protein Data Bank in Europe (PDBe), pays tribute to Michael as a person, describing him as "a dynamo, with an enormous zest for life and science who got along with almost everybody." Gerard also emphasises how, even in recent months, "he was still collecting data himself" and recalls Michael's dismay at a grant funding rejection and how "he simply could not understand how that was possible as it was obviously solid science."
We are indebted to Michael for his help and support to the wwPDB and to the structural biology community as a whole. As Helen Berman puts it: "Michael was truly unique in his contributions to structural biology and we will be forever grateful for all he did."
 wwPDB AC in 2012
wwPDB AC in 2012
04/16/2019
Announcement of mandatory mmCIF submission published in Acta Cryst D
A new publication in Acta Cryst D announces the mandatory submission of PDBx/mmCIF to the PDB for crystallographic depositions. The authorship includes representatives from the major crystallographic structure refinement software packages, in addition to all wwPDB partners.
We are pleased to share our publication “Announcing mandatory submission of PDBx/mmCIF format files for crystallographic depositions to the Protein Data Bank (PDB)”, published in Acta Cryst D. This publication emphasises that PDBx/mmCIF will be the only format accepted for deposition of PDB structures resulting from macromolecular crystallography (MX), including X-ray, neutron, fiber, and electron diffraction methods via OneDep starting July 1st 2019.
Refmac (CCP4), Phenix.refine, and Buster programs can now output PDBx/mmCIF formatted files. Inclusion of representatives from each these softwares within the authorship of the publication highlights the commitment of the crystallographic software community to the PDBx/mmCIF data model. The deposition of PDBx/mmCIF format files will improve the efficiency of the deposition process and enhance validation through capture of the more extensive experimental metadata supported by PDBx/mmCIF, compared to the legacy PDB format.
More information on outputting and preparing PDBx/mmCIF format files for deposition can be found on the wwPDB website.
03/19/2019
The PDB Archive Reaches a Significant Milestone
With this week's update, the PDB archive has passed the milestone of 150,000 entries, and now contains a total of 150,145.
Established in 1971, this central, public archive has reached this milestone thanks to the efforts of structural biologists throughout the world who collectively contribute a wealth of experimentally-determined protein and nucleic acid structure data, which is made available to researchers all around the world, across many different disciplines.
Four wwPDB data centers support online access to three-dimensional structures of biological macromolecules that help researchers understand many facets of biomedicine, agriculture, and ecology, from protein synthesis to health and disease to biological energy. The archive is large, containing more than 1.9 million files related to these PDB entries and requiring more than 512 gigabytes of storage.
The archive reached the landmark of 100,000 entries in 2014, the International Year of Crystallography. Since that record was set, the PDB continued to grow rapidly, both in number of deposited structures and in the complexity of the data. This growth has been supported by the launch of OneDep, a common global system for deposition, validation, and biocuration of PDB data for supported experimental methods. The OneDep system and the underlying PDBx/mmCIF archive format enable the PDB archive to adapt over time to meet the challenges posed by developments in structural biology. More than 41,000 structures that have been deposited, annotated, and validated using OneDep have now been released into the PDB archive, with many more entries updated to ensure consistency of the archive.
With this week's regular update, the PDB welcomes 262 new structures into the archive. These structures join others vital to research and education in fundamental biology, biomedicine, and bioenergy. Since its inception, the size of the archive has increased tenfold roughly every 10-15 years: the PDB reached 100 released entries in 1982, 1000 entries in 1993, and 10,000 in the year 2000. Now that the 150,000th is made available, more than half of the archive has been released in the past ten years.
The scientific community eagerly awaits the next 150,000 structures and the invaluable knowledge these new data will bring. However, the increasing number, size and complexity of biological data being deposited in the PDB and the emergence of hybrid structure determination methods constitute major challenges for the management and representation of structural data. wwPDB will continue to work with the community to meet these challenges and ensure that the archive maintains the highest possible standards of quality, integrity, and consistency.
Development and future of the PDB archive and wwPDB organization is described in the new reference publication for the PDB archive: Protein Data Bank: the single global archive for 3D macromolecular structure data (Nucleic Acids Res., 2019) and many other papers, including Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive (Methods in Molecular Biology, 2017), How community has shaped the Protein Data Bank (Structure, 2013), and Creating a Community Resource for Protein Science (Protein Science, 2012). A full list is available.
02/19/2019
Mandatory PDBx/mmCIF format files submission for MX depositions
Submission of PDBx/mmCIF format files for crystallographic depositions to the PDB will be mandatory from July 1st 2019 onward. PDB format files will no longer be accepted for deposition of structures solved by MX techniques.
PDBx/mmCIF will be the only format accepted for deposition of PDB structures resulting from macromolecular crystallography (MX), including X-ray, neutron, fiber, and electron diffraction methods via OneDep starting July 1st 2019. The deposition of PDBx/mmCIF format files will improve the efficiency of the deposition process and enhance validation through capture of the more extensive experimental metadata supported by PDBx/mmCIF, compared to the legacy PDB format. PDB entries with 100,000 or more atoms, and those with multiple character chain IDs are already not supported by the legacy PDB format. In addition, by 2021, we anticipate the PDB Chemical Component Identifier will need to be extended beyond three characters, which will necessarily result in full retirement of files in the PDB Core Archive that utilize the legacy PDB format.
Refmac, Phenix.refine, and Buster programs can now output PDBx/mmCIF formatted files. For users of other structure determination/refinement software packages, the wwPDB provides stand-alone and web-based tools to convert legacy PDB format files into PDBx/mmCIF format: pdb_extract and MAXIT. More information on outputting and preparing PDBx/mmCIF format files for deposition can be found on the wwPDB website.
The PDBx/mmCIF Working Group has committed to the PDBx/mmCIF data model. PDBx/mmCIF is also supported by visualization software applications, including Jmol/JSMol, LiteMol, Chimera, OpenRasMol, CCP4MG, COOT, PyMOL, VMD, MolMil, and NGL. In addition, other data resources, such as the Protein Model Portal and SASBDB, have adopted and extended the PDBx/mmCIF framework for data representation.
If you have any queries or comments regarding these changes, please contact the wwPDB consortium via deposit-help@mail.wwpdb.org.
02/13/2019
Conclusion of ORI Misconduct Review Results in Obsoletion of PDB Structures, Paper Retractions
In 2018, The Office of Research Integrity (ORI) of the U.S. Department of Health and Human Services announced their final Research Misconduct Finding in the case of H.M. Krishna Murthy. It was found that Murthy reported falsified and/or fabricated research in 10 journal publications and 12 corresponding PDB structures. While the ORI was gathering and evaluating evidence in this case, 5 Murthy structures in the PDB were obsoleted in accord with wwPDB policies, in response to retraction of 4 journal publications. Following a formal request from ORI, received on April 23rd 2018, the remaining 7 Murthy structures in the PDB were obsoleted, again in accord with wwPDB policies. ORI conduct within its investigations is designed to ensure due process for individuals accused of research misconduct, and strict confidentiality is maintained throughout. Only (the final?) findings of research misconduct are made public.
In December 2009, the University of Alabama at Birmingham (UAB) announced that it planned to retract 12 PDB entries and 10 related publications authored by H.M. Krishna Murthy, in his capacity as Principal Investigator and UAB employee. Following wwPDB review of structures at the request of UAB and in accord with wwPDB policy, 5 of the structures were obsoleted upon retraction of the related publications by the journals. Following wwPDB review of structures at the request of UAB and in accord with wwPDB policy, the remaining 7 structures were obsoleted upon receipt of the ORI request.
Since that time, the 5 publications associated with the 7 structures have been retracted by the journals. A detailed PDB history of this case is available.
- RETRACTED: Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans (2001) Cell 104, 301-311 doi: 10.1016/S0092-8674(01)00214-8
- Retraction of "Structures of Apolipoprotein A-II and a Lipid-Surrogate Complex Provide Insights into Apolipoprotein-Lipid Interactions" (2018) Biochemistry 57, 6044 doi: 10.1021/acs.biochem.8b00956
- Retraction for Ganesh et al., Structure of vaccinia complement protein in complex with heparin and potential implications for complement regulation (2018) Proc Natl Acad Sci USA 115, E6965 doi: 10.1073/pnas.1806429115
- Retraction of "Structural Basis for Antagonism by Suramin of Heparin Binding to Vaccinia Complement Protein" (2018) Biochemistry 57, 6043 doi: 10.1021/acs.biochem.8b00955
- Retraction for Ajees et al., Crystal structure of human apolipoprotein A-I: Insights into its protective effect against cardiovascular diseases (2018) Proc Natl Acad Sci USA 115, E6966 doi: 10.1073/pnas.1806430115
Structure Validation and the Role of the PDB as an Archival Data Resource
The PDB is an archival resource that stores, annotates, and disseminates structure models and their related experimental data. The wwPDB has convened expert, community-driven Validation Task Forces for X-ray (in 2008), NMR (in 2009), and (in collaboration with the EMDataBank) Cryo-EM (in 2010) to advise on the most suitable criteria to use for validating structure entries (model, experimental data, and fit of model to data) when they are deposited. Recommendations of these validation task forces have been implemented as part of the wwPDB OneDep system for deposition, annotation, and validation of PDB structures.
The results of these wwPDB validation procedures are captured in a report that is provided to depositors and can be transmitted by them to the journal to which the corresponding manuscript is submitted. Availability of such a report greatly facilitates assessment of the reliability of structural data and its interpretation by journal editors and referees alike. The wwPDB has urged journals publishing structural data on biological macromolecules to require submission of the wwPDB validation report together with the manuscript. The continuing mission of the wwPDB partners is to safeguard the integrity and improve the quality of the structural archive, with the support of the international structural biology community.
For additional information, see
01/02/2019
Time-stamped Copies of the PDB Archive Available
A snapshot of the PDB archive (ftp://ftp.wwpdb.org) as of January 1, 2019 has been added to ftp://snapshots.wwpdb.org and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20190101 includes the 147,610 experimentally-determined structure and experimental data available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot is 1,529 GB.